Our group studies mitochondria & energy metabolism and their regulation in immunity, cancer and metabolic disorders. Our research is divided in three major axes:
Research
I. Mitochondrial plasticity in energy
metabolism and immunometabolism
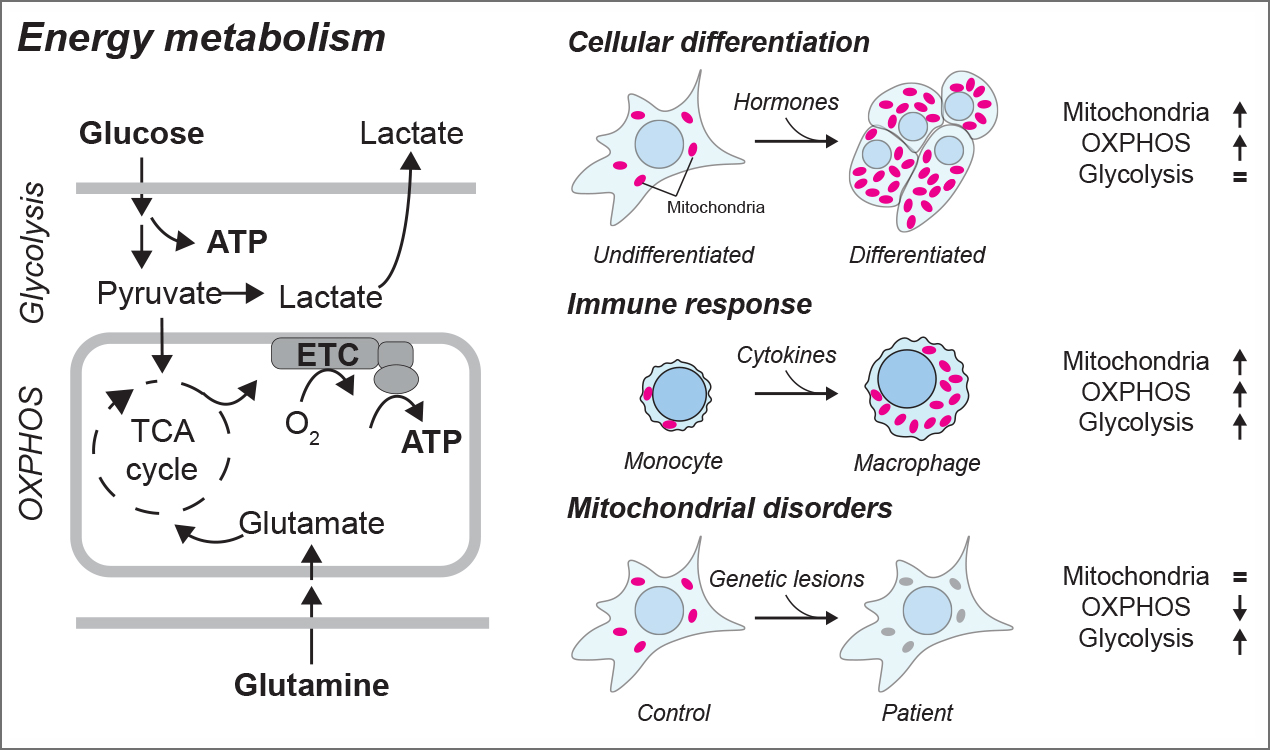
In humans, mitochondrial oxidative phosphorylation (OXPHOS) and glycolysis are the two main pathways for ATP production, or energy metabolism. The reliance on these pathways and the abundance and composition of mitochondria can vary among different tissues, and extreme cases are seen for example during the immune response (immunometabolism). Changes in the OXPHOS/glycolysis balance also underlie conditions such as cancer and metabolic syndromes. Currently, the complete set of molecular mechanisms that regulate and support mitochondrial plasticity and energy metabolism is not fully known. Understanding how these processes function holds potential for diagnosis and therapies by identifying vulnerabilities that could be corrected to promote healthy metabolism and immune function or exploited to inhibit tumor growth.
Figure 1: Energy metabolism pathways and metabolic shifts in physiology and pathology. Left: schematic representation of glycolysis and mitochondrial oxidative phosphorylation (OXPHOS), the two main ATP-generating pathways in human cells. Right: three examples of shifts in metabolic programs and/or in mitochondrial abundance.
Our group uses large-scale systems biology approaches including genetic screening (genome-wide CRISPR/Cas9, ORFeome, RNAi), metabolomics, nutrient screening and quantitative proteomics to discover novel mechanisms involved in mitochondrial plasticity and energy metabolism. Our long-term goals are to:
- Inventory the genetic networks necessary to support energy metabolism. Knowing the identity of these genes will not only advance our understanding of fundamental cellular metabolism but also help prioritize genes for the diagnosis of suspected mitochondrial disorders, a large and heterogenous group of debilitating conditions for which no therapy currently exists.
- Discover novel mechanisms that support dynamic remodeling in mitochondria and energy metabolism.
We use models of cellular differentiation, immune activation and nutrient modulation to identify and manipulate gene expression pathways that control the abundance and composition of mitochondria as well as the expression of metabolic enzymes and transporters. - Study metabolic shifts in pathologies. We aim to apply our findings to understand, and potentially treat, pathologies characterized by metabolic shifts, including mitochondrial disorders, cancers and immune-related disorders.
Representative publications
Salvage of ribose from uridine or RNA supports glycolysis in nutrient-limited conditions.
Skinner OSS*, Blanco-Fernández* J, Goodman RP, Kawakami A, Shen H, Kemény LV, Joesch-Cohen L, Rees MG, Roth JA, Fisher DE, Mootha VK, Jourdain AA. (*co-first authors)
Nature Metabolism 2023
Loss of LUC7L2 and U1 snRNP subunits shifts energy metabolism from glycolysis to OXPHOS.
Jourdain AA, Begg BE, Mick E, Shah H, Calvo SE, Skinner OS, Sharma R, Blue SM, Yeo GW, Burge CB, Mootha VK.
Molecular Cell 2021
A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation.
Arroyo JD*, Jourdain AA*, Calvo SE, Ballarano CA, Doench JG, Root DE, Mootha VK. (*co-first authors)
Cell Metabolism 2016
II. Mitochondrial gene expression and mitochondrial disorders
Mitochondrial disorders are a heterogeneous group of conditions caused by defects in the mitochondria, which are small organelles that play a crucial role in cell metabolism and energy production. These disorders can be inherited or can occur spontaneously and are caused by mutations in either the nuclear DNA or the mitochondrial DNA. Common symptoms of mitochondrial disorders include muscle weakness, developmental delays, intellectual disability, vision or hearing problems, and multi-organ dysfunction. There are currently no successful treatments for most mitochondrial disorders.
The mitochondrial DNA (mtDNA) is a small circular molecule that contains only 37 genes, 13 of which are core subunits of the respiratory chain. It is not fully understood how mtDNA is expressed or how it responds to the environment. This is an important area of investigation because many of the genetic mutations that cause mitochondrial disorders are related to mtDNA. Additionally, it has been discovered that mtDNA and its transcripts are often released from mitochondria into the cytosol, where they can activate pathways of innate immunity, contribute to inflammation, and influence the antiviral response through mechanisms that remain elusive.
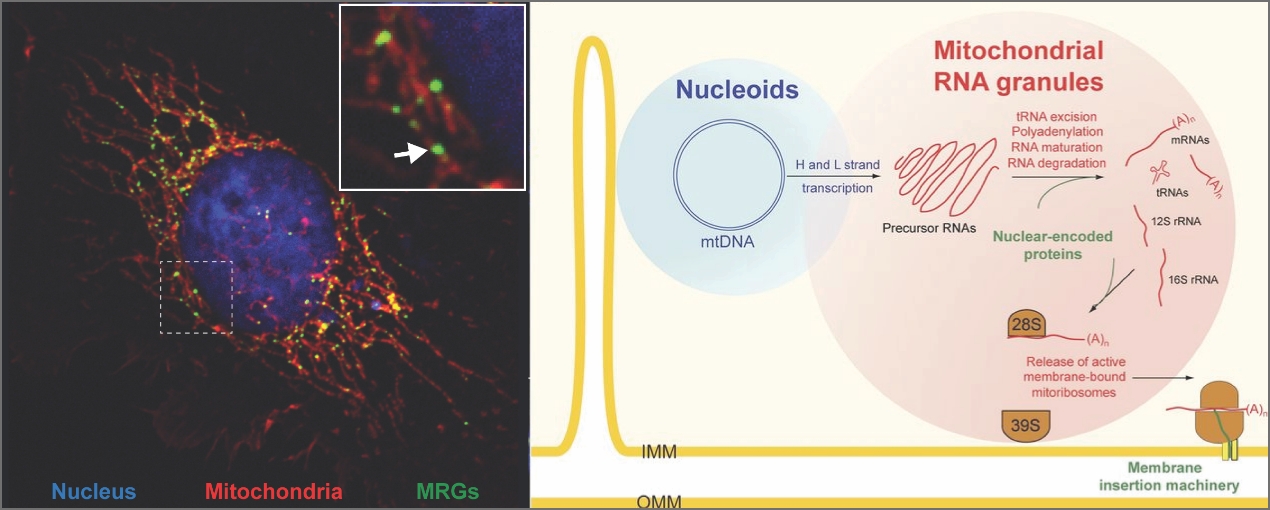
A few years ago, we discovered the existence of «mitochondrial RNA granules» (MRGs) (Fig. 2). We and others have provided evidence that these structures play a vital role in mtDNA expression by serving as post-transcriptional factories for newly-synthesized mitochondrial transcripts. There are still many exciting questions to be answered about the composition and function of these intra-mitochondrial structures.
Figure 2: Mitochondrial gene expression and mitochondrial RNA granules (MRGs). Left: a human cell in which the nucleus, the mitochondrial network and the MRGs (arrow) were labelled with fluorescent dyes. Right: model of mitochondrial gene expression in MRGs. Adapted from Jourdain et al., J. Cell. Biol. 2016.
Our laboratory uses genomics and super-resolution microscopy to characterize the mechanisms of mitochondrial gene expression and their dysfunction in mitochondrial disorders, with a particular emphasis on mitochondrial RNA granules. Our long-term goals are to:
- Discover novel genes involved in mitochondrial gene expression, using large-scale genomics and biochemical technologies, and study their roles in mitochondrial pathologies.
- Characterize the genetic-biophysical properties of mitochondrial RNA granules, by quantitatively studying these structures using simultaneous super-resolution microscopy and genome perturbation.
- Understand how mitochondrial gene expression is impacted by the metabolic state of the cell, including in innate immunity, by combining state of the art gene expression analysis, nutrient source variation and metabolic studies in immune cells (immunometabolism).
The FASTK family of proteins: emerging regulators of mitochondrial RNA biology.
Jourdain AA, Popow J, de la Fuente MA, Martinou JC, Anderson P, Simarro M.
Nucleic Acids Research 2017
Mitochondrial RNA granules: Compartmentalizing mitochondrial gene expression.
Jourdain AA, Boehm E, Maundrell K, Martinou JC.
Journal of Cell Biology 2016
A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function.
Jourdain AA, Koppen M, Rodley CD, Maundrell K, Gueguen N, Reynier P, Guaras AM, Enriquez JA, Anderson P, Simarro M and Martinou JC.
Cell Reports 2015
GRSF1 regulates RNA processing in mitochondrial RNA granules.
Jourdain AA, Koppen M, Wydro M., Rodley CD, Lightowlers RN, Chrzanowska-Lightowlers ZM, Martinou JC.
Cell Metabolism 2013
III. Nucleotide metabolism
Nucleotides are the building blocks of nucleic acids (DNA and RNA), and each nucleotide is made up of a five-carbon sugar molecule, a phosphate group, and a nitrogenous base. In addition to their role in nucleic acids, nucleotides also play important roles in many other biological processes, including energy metabolism (ATP, NADH), cell signaling, and regulation of gene expression. While the synthesis of purine nucleotides occurs exclusively in the cytosol, a crucial step in the synthesis of pyrimidine nucleotides occurs in mitochondria, connecting nucleotide synthesis and OXPHOS.
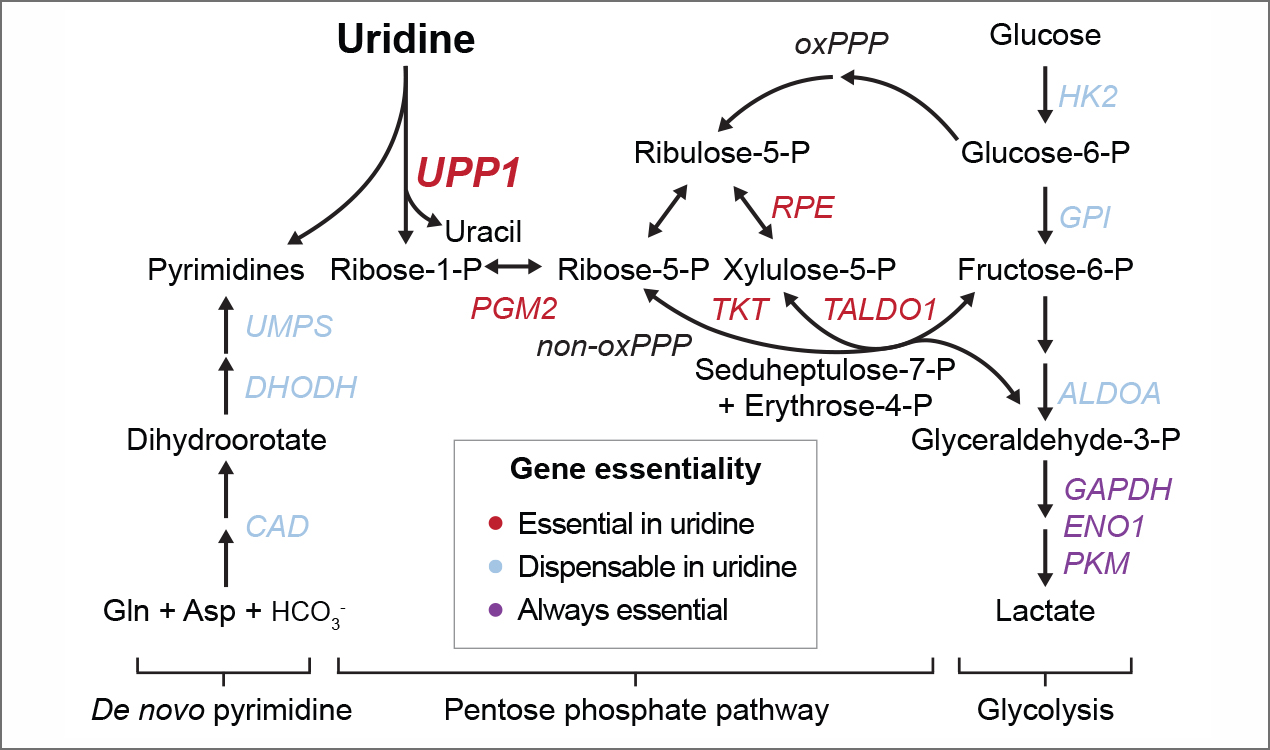
We recently discovered that uridine and RNA can serve as substrates for glycolysis, a metabolic pathway in which those nucleosides and nucleic acids can potentially serve as alternative sources of energy. By using a combination of genetic screens, metabolic tracers, and in vivo experiments, we have demonstrated that glycolysis from uridine and RNA relies on the activity of the uridine phosphorylase genes UPP1 and UPP2, and the pentose phosphate pathway to obtain high-energy carbon from the sugar molecule in uridine (Fig. 3). Glycolysis from uridine and RNA appears to be widespread, and we have confirmed its activity in cancer cells, primary macrophages, and in mice in vivo.
Figure 3: Pyrimidine synthesis and catabolism for energy production. Left: Pyrimidine nucleotides are synthesized de novo from amino acids and bicarbonate, a process that involves mitochondria (not shown). Center and right: Uridine degradation for energy production. Carbons from the sugar of uridine are shuttled through the pentose phosphate pathway to glycolysis.
We are currently using genetic screening, high-throughput drug screening and physiological models to better understand, and modulate, nucleotide metabolism. Our goals are to:
- Explore the occurrence of glycolysis from uridine and RNA in the organism and its physiological regulation in animal models and during dietary modulation.
- Discover small molecules inhibitors of glycolysis from uridine and RNA, by performing high-throughput drug screening. Those inhibitors have a strong potential for oncology, immune modulation or for the treatment of metabolic disorders such as obesity.
- Identify novel genes and factors involved in de novo pyrimidine synthesis using genetic screens and metabolomics.
Representative publication
Salvage of ribose from uridine or RNA supports glycolysis in nutrient-limited conditions.
Skinner OSS*, Blanco-Fernández* J, Goodman RP, Kawakami A, Shen H, Kemény LV, Joesch-Cohen L, Rees MG, Roth JA, Fisher DE, Mootha VK, Jourdain AA. (*co-first authors)
Nature Metabolism 2023
Funding
Our research is funded by the Swiss National Science Foundation (SNSF), the foundation Pierre-Mercier pour la Science, the European Molecular Biology Organization (EMBO), the Swiss Federation for Research on Muscular Disorders (FSRMM), the Olga Mayenfisch Stiftung, and the University of Lausanne.